Tophat is a splicing aware aligner so we can map transcripts to the genome. If theres no index for your organism its easy to build one yourself.
Introduction To Bulk Rnaseq Analysis Bioinformatics Documentation
RNA-Seq and TopHatThis video uses animation and the GEP UCSC Genome Browser to illustrate how RNA-Seq data are displayed in the Genome Browser and how TopHat.
. The following tutorial demonstrates how to use TopHat-Fusion to identify fusion genes including three known fusions BCAS4-BCAS3 ARFGEF2-SULF2 RPS6KB1-TMEM49. This video uses animation and the GEP UCSC Genome Browser to illustrate how RNA-Seq data are displayed in the Genome Browser and how TopHat predicts splice j. TopHat-Fusion consists of two sub-programs tophat and tophat-fusion-post.
Bowtie1 is an ultrafast memory-effi cient aligner for large sets of short reads. The user ID is normally your Cornell NetID. At the very end we can compare these results to the results we got from mapping directly to the.
Some of the applications used in RNA sequencing analysis are the following. Configure the package specifying the install path and the library dependencies as needed. The Bowtie site provides pre-built indices for human mouse fruit fly and others.
Using a breast cancer cell MCF7 RNA-Seq data from Edgren et al Genome Biology 2011. RNA-Seq Tutorials Tutorial 1 RNA-Seq experiment design and analysis Instruction on individual software will be provided in other tutorials Tutorial 2 Advanced RNA-Seq Analysis topics Hands-on tutorials Analyzing human and potato RNA-Seq data using Tophat and Cufflinks in Galaxy. Everyone should have a BioHPC account to access the computer.
Alignment with TOPHAT Part 1. RNA-Seq Tutorials Tutorial 1 RNA-Seq experiment design and analysis Instruction on individual software will be provided in other tutorials Tutorial 2 Hands-on using TopHat and Cufflinks in Galaxy Tutorial 3 Advanced RNA-Seq Analysis topics. RNA-seq Read Mapping with TOPHAT and STAR.
Using TophatCufflinks to analyze RNAseq data. Version 120 includes some important bug fixes as well as a major new feature. Some of the applications used in RNA sequencing analysis are the following.
TopHat2 uses using Bowtie to align RNA-Seq reads to mammalian-sized genomes and then analyzes the mapping results to identify splice junctions between exons. Find out the name of the computer that has been reserved for you httpscbsutccornelleduwwmachinesaspxi88. In this tutorial well map reads from an RNA-seq study in Drosophila melanogaster to the reference genome using tophat.
We recommend that you watch the video Aligning RNA-seq reads to reference genome instead which covers t. Everyone should have a BioHPC account to access the computer. Introduce the types of files typically used in RNA-seq analysis.
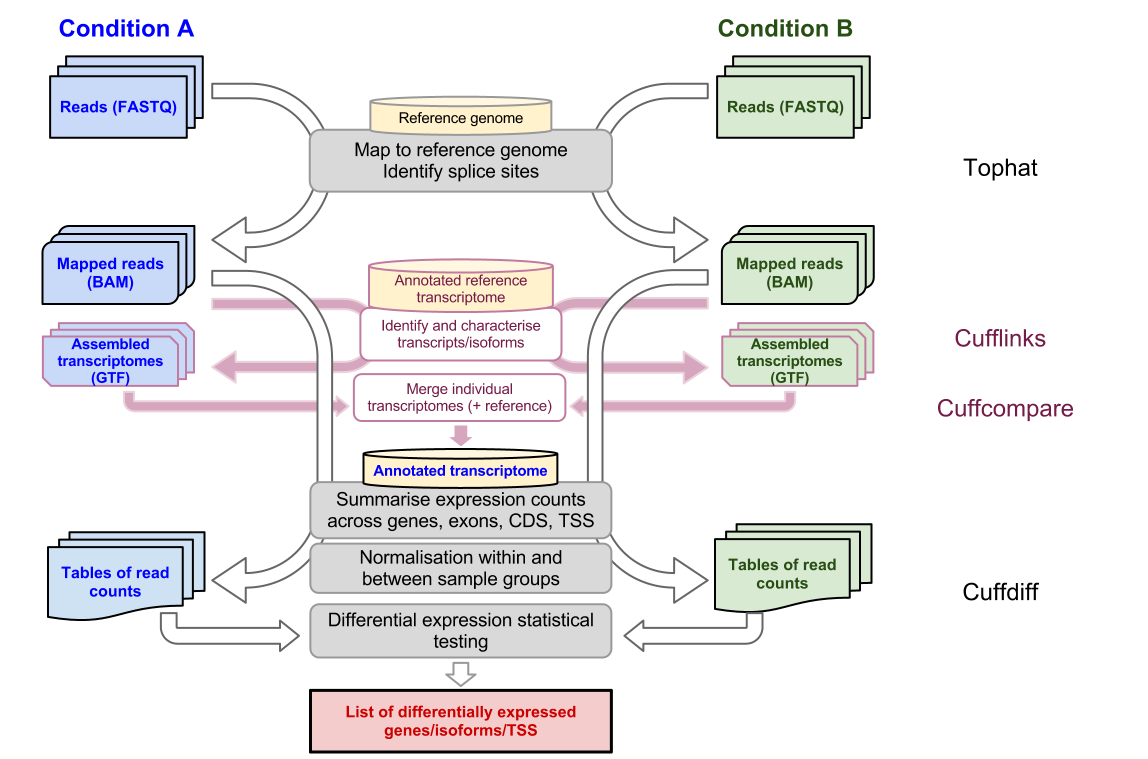
Our input data for this tutorial will be raw RNA-seq reads from two experimental conditions and we will output a list of differentially expressed genes identified to be statistically significant. Mapping to a reference genome with tophat and counting with HT-seq. TopHat-Fusion consists of two sub-programs tophat and tophat-fusion-post.
If you have Bowtie 2 installed and want to use it with Tophat v20 or later you must create Bowtie 2 indexes for your. To find junctions with TopHat youll first need to install a Bowtie index for the organism in your RNA-Seq experiment. Bowtie1 is an ultrafast memory-effi cient aligner for large sets of short reads.
In this tutorial well use some sample data from a project we did on flies Drosophila melanogaster to illustrate how you can use RNA-seq data to look for differentially expressed genes. Find out the name of the computer that has been reserved for you httpcbsutccornelledulabdocworkstations_rnaseq_2014_04_04htm. One of CBSU BioHPC Lab workstations has been allocated for your workshop exercise.
TopHat is a collaborative effort between the University of Maryland Center for Bioinformatics and Computational Biology and the University of California Berkeley Departments of Mathematics. Well try a few different approaches to see whether different tools give similar results. The user ID is normally your Cornell NetID.
The following tutorial demonstrates how to use TopHat-Fusion to identify fusion genes including three known fusions BCAS4-BCAS3 ARFGEF2-SULF2 RPS6KB1-TMEM49. Align RNA-seq reads with Tophat. Using a breast cancer cell MCF7 RNA-Seq data from Edgren et al Genome Biology 2011.
Httpcbsutccornelleduww1sessionaspxwid9sid12 Please consult the PDF file with instructions on how to access and use the Lab workstations for the. This is quite different conceptually to mapping to the transcriptome directly. Thanks to the efforts of Ryan Kelley from Illumina TopHat now supports detection of insertions and deletions using RNA-Seq data.
Prepare the working directory. Prepare the working directory. To install TopHat from source package unpack the tarball and change directory to the package directory as follows.
It aligns RNA-Seq reads to mammalian-sized genomes using the ultra high-throughput short read aligner Bowtie and then analyzes the mapping results to identify splice junctions between exons. TopHat will report reads aligned across discovered indels by taking advantage of I and D CIGAR operators as specified by the SAM format. The allocations are listed on the workshop exercise web page.
This tutorial from 2017 covers the TopHat aligner. In this tutorial we will. TopHat2 uses using Bowtie to align RNA-Seq reads to mammalian-sized genomes and then analyzes the mapping results to identify splice junctions between exons.
Basic Analyses With Tophat Cufflinks Rnaseq Tutorial 1 Documentation

Reference Based Rnaseq Data Analysis Long
The Cufflinks Rna Seq Workflow
Tophat Cufflinks Command Pipeline

Rna Seq Alignment And Visualization Youtube
Incorporating Rnaseq Tophat To Augustus Computational Biology

0 comments
Post a Comment